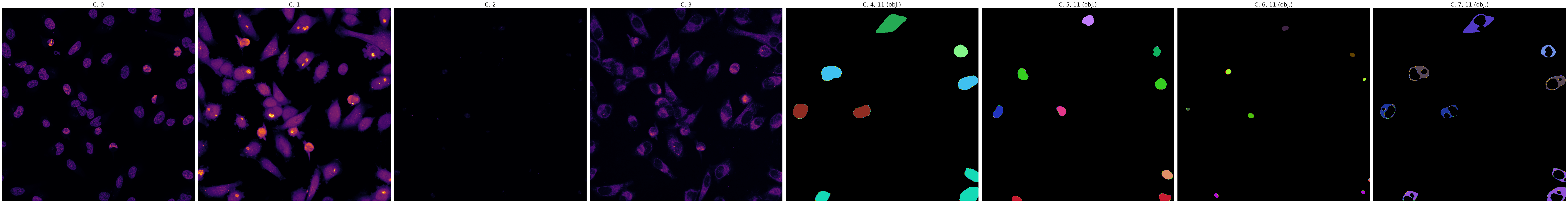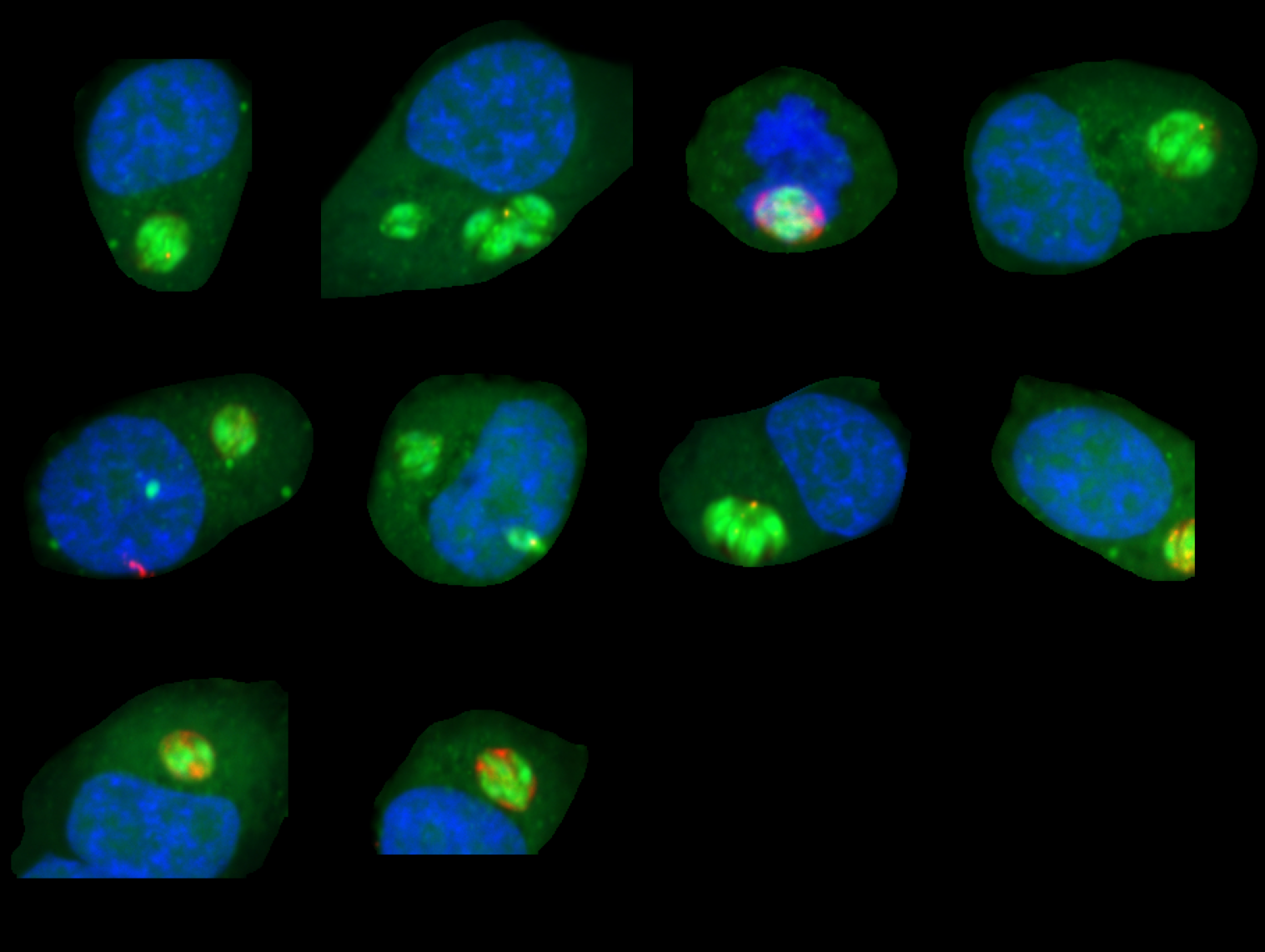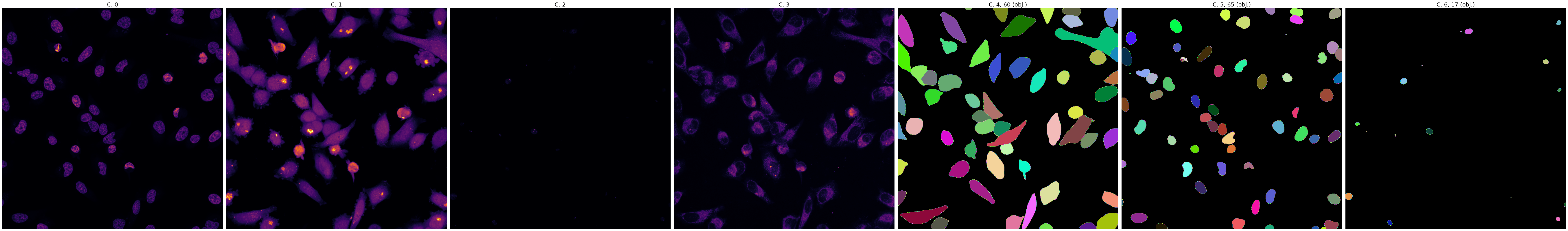
This slide demonstrates the initial mask generation using the Cellpose 'cyto' model. Note the high signal-to-noise
ratio achieved by preprocessing with background removal and normalization enabled.
from spacr.measure import measure_crop
%matplotlib inline
settings = {
'src':'/home/olafsson/datasets/plate1/test/merged',
'channels':[0,1,2,3],
'cell_mask_dim':4,
'cell_min_size':2000,
'nucleus_mask_dim':5,
'nucleus_min_size':1000,
'pathogen_mask_dim':6,
'pathogen_min_size':400,
'cytoplasm_min_size':0,
'save_png':True,
'crop_mode':['cell'],
'use_bounding_box':False,
'png_size':[[224,224]],
'normalize':False,
'png_dims':[0,1,2],
'normalize_by':'png',
'save_measurements':True,
'plot':True,
'plot_filtration':False,
'uninfected':False,
'test_mode':False,
'test_nr':10
}