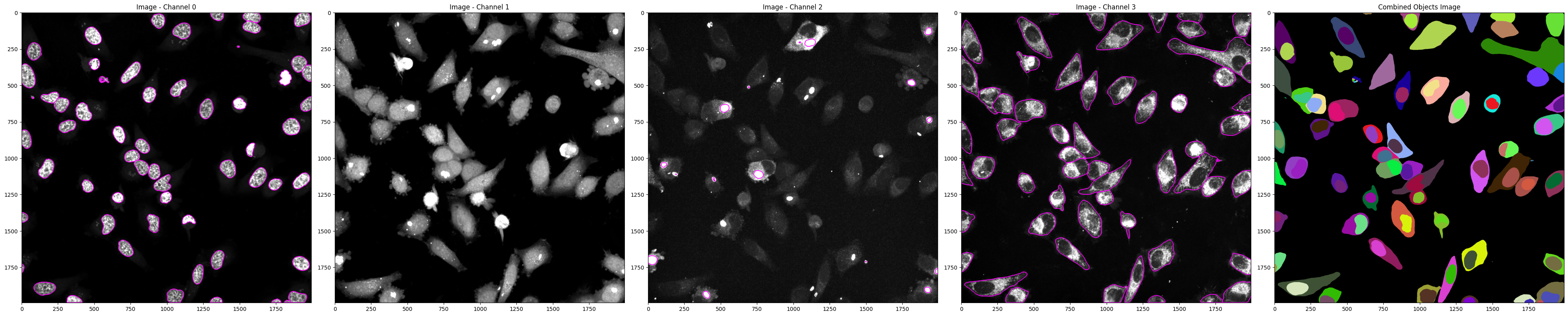
from spacr.core import preprocess_generate_masks
%matplotlib inline
settings = {
'src':'/home/olafsson/datasets/plate1',
'metadata_type':'cellvoyager',
'custom_regex':None,
'experiment':'screen',
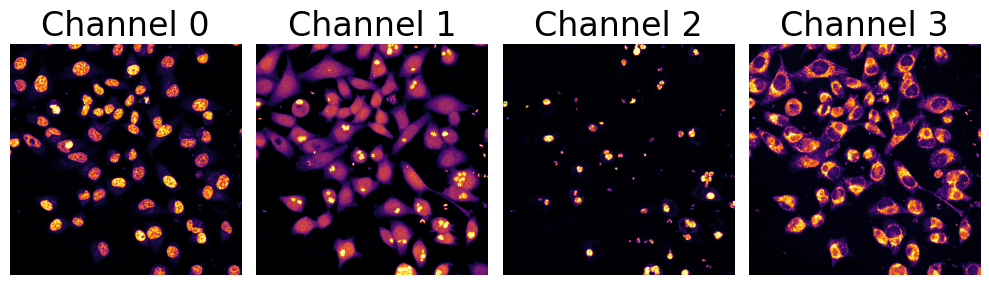
'channels':[0,1,2,3],
'cell_channel':3,
'cell_background':100,
'cell_Signal_to_noise':10,
'cell_CP_prob':-1,
'remove_background_cell':False,
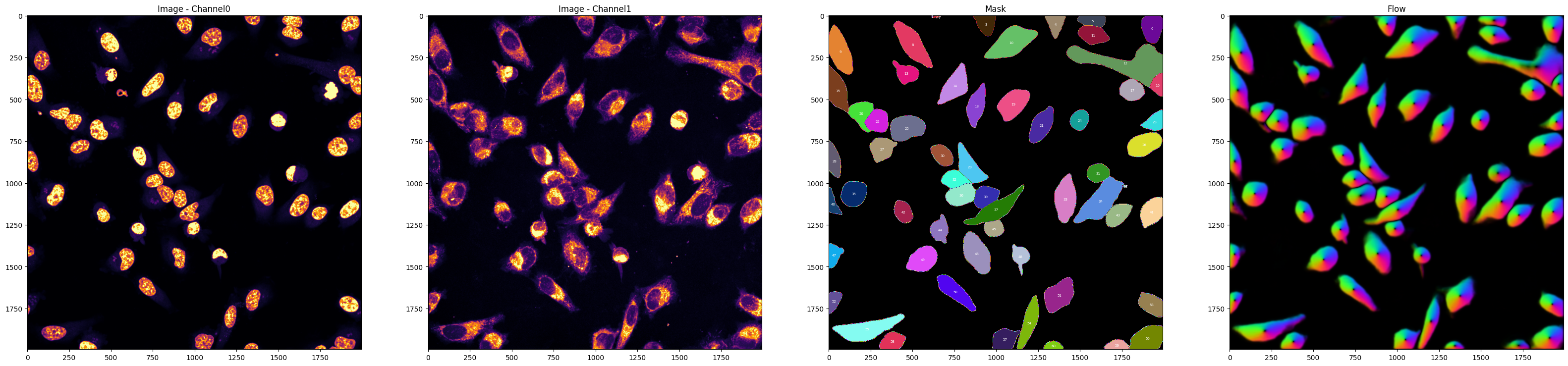
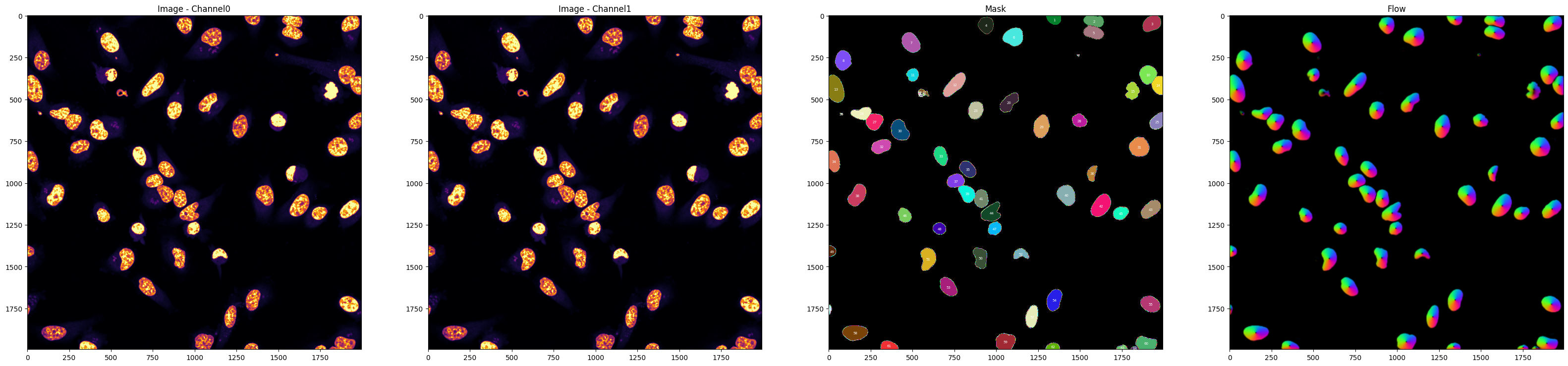
'nucleus_channel':0,
'nucleus_background':200,
'nucleus_Signal_to_noise':5,
'nucleus_CP_prob':0,
'remove_background_nucleus':False,
'pathogen_model':None,
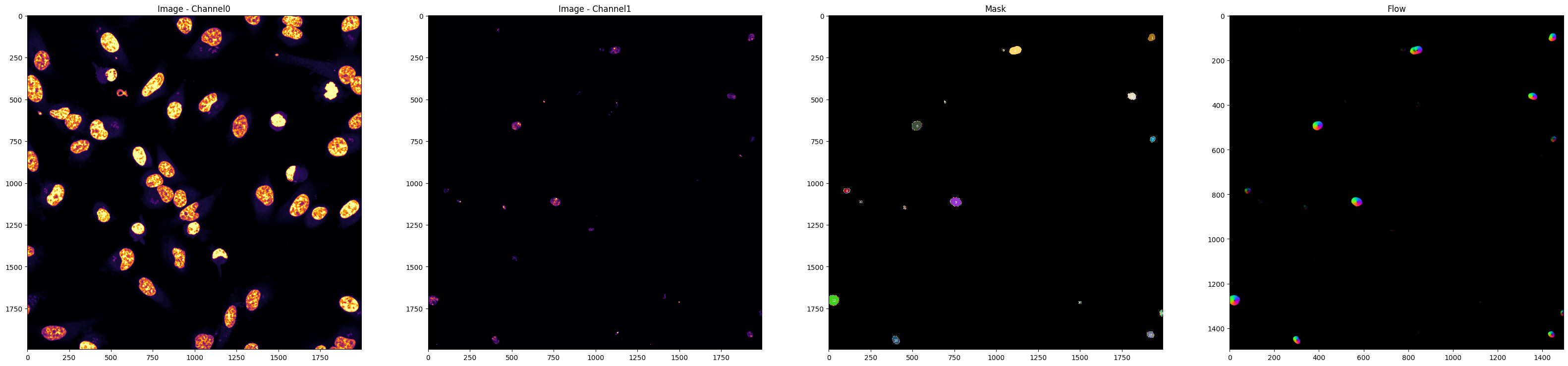
'pathogen_channel':2,
'pathogen_background':400,
'pathogen_Signal_to_noise':5,
'pathogen_CP_prob':-2,
'remove_background_pathogen':True,
'consolidate':False,
'magnification':40,
'save':True,
'preprocess':True,
'masks':True,
'batch_size':50,
'filter':False,
'merge_pathogens':False,
'plot':True,
'adjust_cells':True,
'test_mode':True,
'test_images':10,
'random_test':True
}